From decades of detailed anatomical studies, scientists have learned about the close relationship between anatomy and function. However, while we know a lot about which regions of the brain connect to one another, the features of brain structure essential to our understanding of function are currently unknown.
Thanks to the advent of new technologies that enable researchers to visualize different cell types in the brain, neuroscientists are trying to piece together how all these cells form larger structures with specific functions.
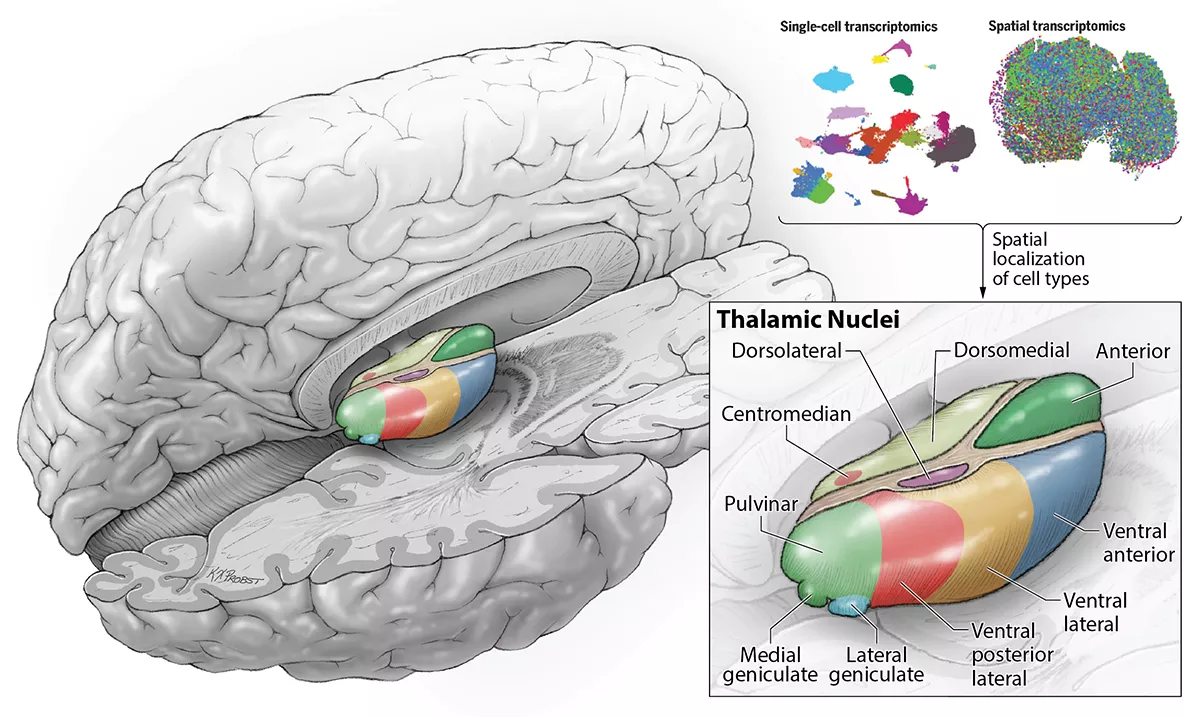
Researchers at UC San Francisco have now identified the various cell types found in the thalamus, a region of the brain that serves as a central node for relaying sensory and motor information from the body to other areas of the brain. Their findings offer the most comprehensive view to date of when distinct clusters of neurons, known as nuclei, form during development.
This research is part of a series of studies published in Science, Science Advances, and Science Translational Medicine, that highlight the latest efforts of the National Institutes of Health’s BRAIN Initiative Cell Census Network (BICCN) — a project launched in 2017 to develop reference maps of all the cell types in the brains of mice, primates, and humans.
“What is special about the brain is that hundreds of different cell types interact with one another in ways that make the ensembles be greater than the sum of the parts,” said Tomasz Nowakowski, PhD, an associate professor in the Department of Neurological Surgery and the study’s senior author.

In the thalamus, nuclei are easily distinguishable under a microscope, and each of the thalamic nuclei have well-defined connections to other areas of the brain.
The researchers, led by UCSF graduate students Chang Kim and David Shin, used single cell RNA sequencing to classify all the cell types in the developing human thalamus based on the abundance of all their genes. With a technique called spatial transcriptomics, they could also see where the different cells they were identifying were located within the thalamus. These experiments helped them overlay the new information about the molecular features of the cells onto the existing diagrams of brain anatomy.
To their surprise, the scientists found that while some cell types could only be found in certain thalamic nuclei, others were dispersed across many different nuclei with distinct functions.
“These findings suggest that thalamic nuclei are really a mosaic made up of molecularly distinct cell types, with complex patterns that emerge early in development,” Nowakowski, who is also a faculty member in the Departments of Anatomy and Psychiatry and Behavioral Sciences as well as the UCSF Eli and Edythe Broad Center for Regeneration Medicine and Stem Cell Research, said.
Additionally, Nowakowski’s lab identified cells in the developing thalamus from other distant regions of the brain. These populations of cells, he says, seem to be much more abundant in the brains of all primates, including humans, compared to mice.
“This strategy represents one way in which the more complex brains of primates and humans may have evolved,” he said.
Nowakowski and his team now hope to figure out how these cells contribute to more complex cognitive processes.
This cell atlas also helps scientists start to understand how problems in the developing human brain can give rise to neuropsychiatric disorders like autism spectrum disorder and schizophrenia.
Reference:
Kim CN, Shin D, Wang A, Nowakowski TJ. Spatiotemporal molecular dynamics of the developing human thalamus. Science. 2023 Oct 13;382(6667):eadf9941. doi: 10.1126/science.adf9941. Epub 2023 Oct 13. PMID: 37824646.
